
The ontologies of the anatomy track are the NCI Thesaurus describing the human anatomy, published by the National Cancer Institute (NCI), and the Adult Mouse Anatomical Dictionary, which has been developed as part of the Mouse Gene Expression Database project. Both resources are part of the Open Biomedical Ontologies (OBO). A detailed description of the data set has been given in the context of OAEI 2007 and 2008.
As proposed in 2008 the task of automatically generating an alignment has been divided into four subtasks. Task #1 is obligatory for participants of the anatomy track, while task #2, #3 and #4 are optional tasks.
Due to the harmonization of the ontologies applied in the process of generating a reference alignment (see [1] and anatomy OAEI 2007 page), a high number of rather trivial correspondences (61%) can be found by simple string comparison techniques. At the same time, we have a good share of non-trivial correspondences (39%). The partial reference alignment used in subtrack #4 is the union of all trivial correspondences and 54 non-trivial correspondences.
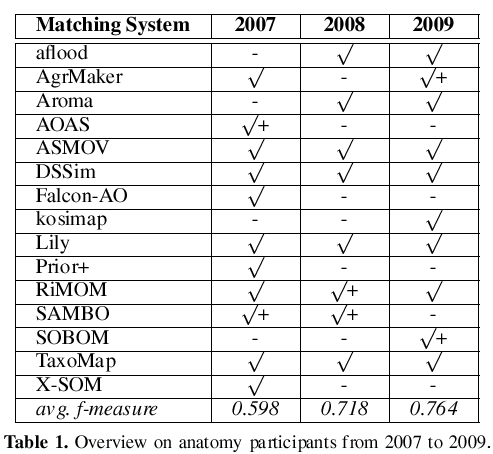
In Table 1 we have marked the participants with an f-measure > with a + symbol. Unfortunately, the top performers of the last two years do not participate this year. In the last row of the table the average of the obtained f-measures is shown. We observe significant improvements over time. However, in each of the three years the top systems generated alignments with f-measure of approx. 0.85. It seems that there is an upper bound which is hard to exceed.
Due to the evaluation process of the OAEI, the submitted alignments have been generated by the participants, who run the respective systems on their own machines. Nevertheless, the resulting runtime measurements provide an approximate basis for a useful comparison. In 2007 we observed significant differences with respect to the stated runtimes. Lily required several days for completing the matching task and more than half of the systems could not match the ontologies in less than one hour. In 2008 we already observed increased runtimes and this years evaluation revealed that only one system still requires more than one hour. The fastest system is aflood (15 sec) followed by Aroma, which requires approximately 1 minute. Notice that aflood is run with a configuration optimized for runtime efficiency in task #1, it requires 4 minutes with a configuration which aims at generating an optimal alignment used for #2, #3, and #4. Detailed information about runtimes can be found in the second column of Table 2.
Table 2 lists the results of the participants in descending order with respect to the f-measure achieved for subtrack #1. In the first two rows we find SOBOM and AgreementMaker. Both systems have very good results and distance themselves from the remaining systems. SOBOM, although participating for the first time, submitted the best result in 2009. The system seems to be optimized for generating a precise alignment, however, the submitted alignment contains also a significant number of non trivial correspondences (see the column Recall+ for subtrack #1).1
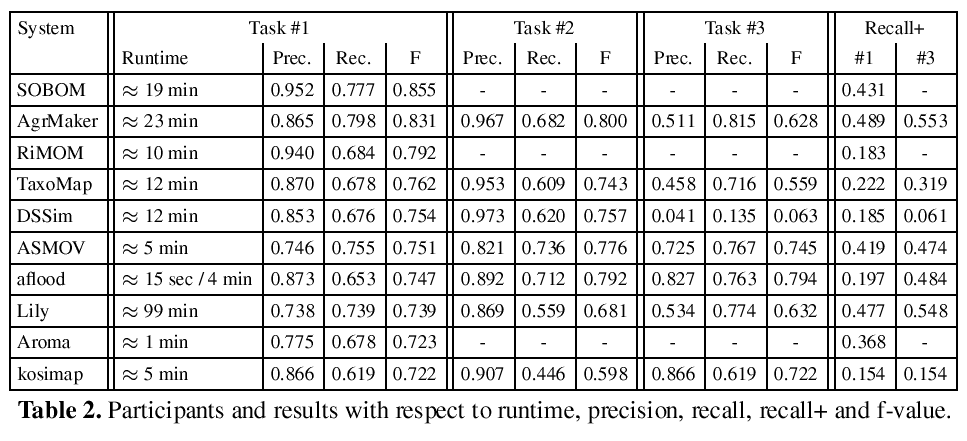
AgreementMaker generates a less precise alignment, but manages to output a higher number of correct correspondences. None of the other systems detected a higher number of non-trivial correspondences for both subtrack #1 and #3 in 2009. However, it cannot top the SAMBO submission of 2008, which is known for its extensive use of biomedical background knowledge.
The RiMOM system is slightly worse with respect to the achieved f-measure compared to its 2008 submission. The precision has been improved, however, this caused a loss of recall and in particular a siginficant loss of recall+. Unfortunately, RiMOM did not particpate in subtask #3, so we cannot make statements about its strength in detecting non-trivial correspondences based on a different configuration.
The systems listed in the following columns achieve similar results with respect to the overall quality of the generated alignments (f-measures between 0.72 and 0.76). However, significant differences can be found in terms of the trade-off between precision and recall. All systems except ASMOV and Lily favor precision over recall. Notice that an f-measure of 0.755 can easily be achieved by constructing a highly precise alignment without detecting any non-trivial correspondences. At the same time it is relatively hard to generate an alignment with an f-measure of 0.755 that favors recall over precision. Thus, the results of ASMOV and Lily have to be interpreted more positive than indicated by the f-measure.
The observation that it is not hard to construct a highly precise alignment with acceptable recall is supported by the results of subtask #2, where we find relatively similar results for all participants. In particular, it turned out that for some systems (ASMOV, DSSim) we measured the best f-measure in track #2. The evaluation results for aflood require some additional exlanations. aflood is run for track #1 with a configuration which results in a significant reduction of the runtime (15 sec), while for track #2 and #3 the system required approximately 4 minutes due to different settings. Therefore, aflood creates better alignments as solutions to subtask #2 and #3.
In 2007 we were surprised by the good performance of the naive label comparison approach. Again, we have to emphasize that this is to a large degree based on the harmonization of the ontologies that has been applied in the context of generating the reference alignment. Nevertheless, the majority of participants was able to top the results of the trivial string matching approach this year.
In the following we refer to an alignment generated for task #1 resp. #4 as A1 resp. A4. This year we have chosen an evaluation strategy that differs from the approach of the last year. We compare A1 ∪ Rp resp. A4 ∪ Rp with the reference alignment R. Thus, we compare the situation where the partial reference alignment is added after the matching process has been conducted against the situation where the partial reference alignment is available as additional resource used within the matching process. The results are presented in Table 3.
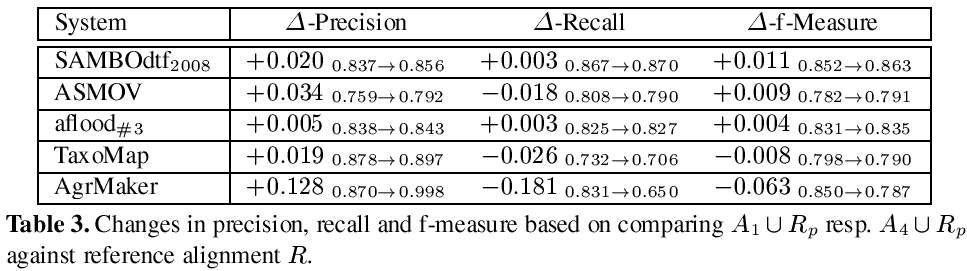
Four systems participated in task #4. These systems were aflood, AgreementMaker, ASMOV and TaxoMap. In Table 3 we additionally added a row that displays the 2008 submission of SAMBOdtf, which had the best results for subtrack #4 in 2008. For aflood we used A3 instead of A1 to allow a fair comparison, due to the fact that A1 was generated with runtime optimization configuration.
A first look at the results shows that all systems use the partial reference alignment to increase the precision of their systems. Most of them them have slightly better values for precision (between 0.5% and 3.4%), only AgreementMaker uses the additional information in a way which has a stronger impact in terms of a significantly increased precision. However, only three correspondences have been found that have not been in the partial reference alignment previously.2 Only SAMBOdtf and aflood profit from the partial reference alignment by a slightly increased recall, while the other systems wrongly filter out some correct correspondences. This might be based on two specifics of the dataset. On the one hand the major part of the reference alignment consists of trivial correspondences easily detectable by string matching algorithms, while the unknown parts share a different characteristic. Any approach which applies machine learning techniques to learn from the partial reference alignment is thus bound to fail. On the other hand parts of the matched ontologies are incomplete with respect to subsumption axioms. As pointed out in [2], the completeness of the structure and the correct use of the structural relations within the ontologies has an important influence on the quality of the results. For these reasons it is extremely hard to use the partial reference alignment in an appropriate way in subtask #4.
Although it is argued that domain related background knowledge is a crucial point in matching biomedical ontologies, the results of 2009 raise some doubts about this issue. While in 2007 and 2008 the competition was clearly dominated by matching systems heavily exploiting background knowledge (UMLS), this years top performer SOBOM uses none of these techniques. However, the strong f-measure of SOBOM is mainly based on high precision. Comparing the alignments generated by SAMBO in 2008 and SOBOM in 2009 it turns out that SAMBO detected 136 correct correspondences not found by SOBOM, while SOBOM finds 36 correct correspondences not detected by SAMBO. Unfortunately, SOBOM did not participate in subtrack #3. Thus, it is hard to assess its capability for detecting non-trivial correspondences. The results of subtask #4 are disappointing at first sight. Since in 2008 this kind of task has been introduced for the first time, we expected better results in 2009. However, it turned out again that only minor positive effects can be achieved. But, as already argued, the task of subtrack #4 is hard and systems with acceptable results in subtrack #4 might obtain good results under better conditions.
The ZIP-file contains all final submissions in the format matcher_tracknumber.rdf in the same folder.
(1) The developer of the TaxoMap system closely missed the final submission deadline and final optimizations of their systems are thus not reflected in the tables presented above. Here are the new scores for their improved submissions referred to as TaxoMapX in the following.
TaxoMapX Prec. Rec. F-meas. Subtask #1 0.866 0.724 0.789 Subtask #2 0.954 0.679 0.793 Subtask #3 0.779 0.742 0.760 and R+ for #1/#3 is 0.287/0.333
As can be seen the results have become better, however, the two leading systems cannot be reached.The following results are available for subtask #4, where we observe some changes. The main tendency did not change in this specific task.
Change
Prec. 0.039 with 0.871 -> 0.910
Rec. -0.048 with 0.757 -> 0.709
F-meas. -0.013 with 0.810 -> 0.797
(2) System AgreementMaker has been referred to as AgrMaker in tables and AgreementMaker in the text.
(3) In a first version of this webpage (and the results paper) we forgot to include the system RiMOM. This has been fixed. The system particpated in subtrack #1 (see Tables 1 and 2) and has been included in the discussion of the evaluation results.
This track is organized by Christian Meilicke and Heiner Stuckenschmidt. If you notice any errors or misleading remarks on this page, directly contact Christian Meilicke (email to christian [at] informatik [.] uni-mannheim [.] de).
1Recall+ is defined as recall restricted to the subset of non trivial correspondences in the reference alignment. A detailed definition can be found in results section at OAEI 2007.
2 Notice that we only take correspondences between anatomical concepts into account.
[1] Bodenreider O. and Hayamizu T. and Ringwald M. and De Coronado S. and Zhang S.: Of Mice and Men: Aligning Mouse and Human Anatomies. Proceedings of the American Medical Informatics Association (AIMA) Annual Symposium, 2005.
[2] Lambrix P. and Liu Q.: Using partial reference alignments to align ontologies. Proceedings of the 6th European Semantic Web Conference. Heraklion, Greece, 2009.